Rust, Puccinia
Trait Symbol: Leaf rust
Page Contributors: PeanutBase (Jugpreet Singh)
Major Information Sources:
- Rajeev K. Varshney, Manish K. Pandey, Pasupuleti Janila, Shyam N. Nigam, Harikishan Sudini, M.V.C. Gowda, Manda Sriswathi, T. Radhakrishnan, Surendra S. Manohar, Patne Nagesh. Marker-assisted introgression of a QTL region to improve rust resistance in three elite and popular varieties of peanut (Arachis hypogaea L.). Theoretical and Applied Genetics (2014) 127(8):1771-1781.
- Suvendu Mondal, Poonam Hande, Anand M. Badigannavar. Identification of Transposable Element Markers for a Rust (Puccinia arachidis Speg.) Resistance Gene in Cultivated Peanut. Journal of Phytopathology (2014) 162: 548-552
• IPAHM103 • GM1536 • GM2301 • GM2079
• TE 360 • TE 498
• SSR_GO340445 • SSR_HO115759
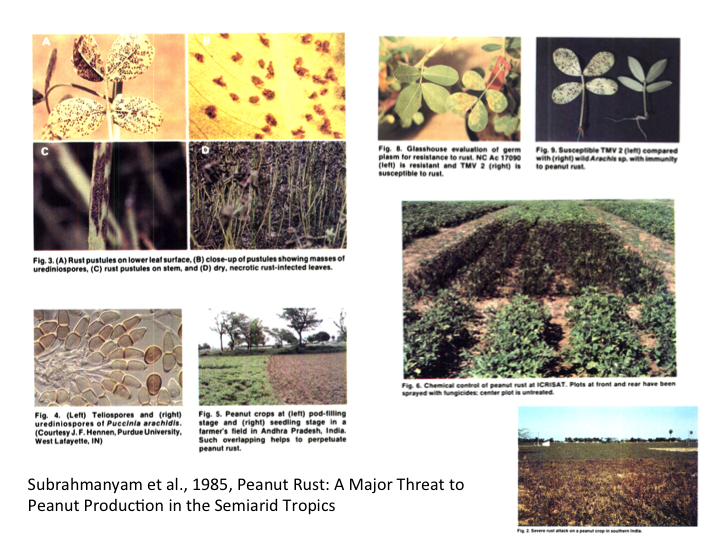
Background Information: Leaf rust, caused by Puccinia arachidis, is a widespread disease in peanut that leads to severe yield losses in almost all peanut growing countries. Together with late leaf spot, leaf rust causes signifcant decrease in productivity and fodder quality in peanut.
Trait Details: Trait synonyms: Fungal disease resistance, Leaf Rust (Puccinia arachidis Speg)
Peanut leaf rust is caused by Puccinia arachidis. The synonym terms used for this fungus are Bullaria arachidis (Speg.) Arthur & Mains, (1922) and Uromyces arachidis Henn. (1896) [Wikipedia]. The term rust is used for both the disease and the causative fungus (Sillero et al., 2006).
The disease symptoms can be recognized by the appearance of orange pustules on the lower surface of the leaves. The rupturing of these pustules expose masses of reddish brown urediniospores which leads to the spread of infection to the neighboring plants. In highly susceptible genotypes, secondary pustules might surround the original pustules. Later, pustules may form on the upper surface corresponding to the position on the lower surface of leaf. These pustules develop on all the plant parts except flowers. Pustules are usually circular in shape that ranges from 0.5 to 1.4mm in diameter. Rust infection lead to dried and necrotic leaves which stay intact with the plant. The disease is usually spread through urediniospores by air, infected crop debris like surface contaminated pods, seeds or dried leaves. However, spread through seed borne pathogen or germplasm exchange is not evident. Leaf rust can cause infection to plants at any development age. Also, warm and humid weather can facilitate the spread of this fungus. Under favorable circumstances, leaf rust can spread to the entire field and can cause desiccation of all the plants. Early season rust infection may lead to significant yield losses as compared to late season rust epidemics (Sillero et al., 2006; Rashid & Bernier, 1991). In essence, severe disease infection can lead to approximately 50% of the yield losses.

Markers for MAS:
| Marker | IPAHM103 | GM1536 | GM2301 | GM2079 |
|---|---|---|---|---|
| Source | Varshney, Pandey et al., 2014 | Varshney, Pandey et al., 2014 | Varshney, Pandey et al., 2014 | Varshney, Pandey et al., 2014 |
| Comments | "The marker-assisted backcrossing approach employed a total of four markers including one dominant (IPAHM103) and three co-dominant (GM2079, GM1536, GM2301) markers present in the QTL region." (Varshney, Pandey et al., 2014) | |||
| Type | SSR | |||
| Repeat | NULL | |||
| Forward primer | GCATTCACCACCATAGTCCA | AAAGCCCTGAAAAGAAAGCAG | GTAACCACAGCTGGCATGAAC | GGCCAAGGAGAAGAAGAAAGA |
| Reverse primer | TCCTCTGACTTTCCTCCATCA | TATGCATTTGCAGGTTCTGGT | TCTTCAAGAACCCACCAACAC | GAAGGAGTAGTGGTGCTGCTG |
| Size (bp) in susceptible lines | 130 | 482 | 136 | 436 |
| Size (bp) in resistant lines | 154 | 473 | 127 | 416 |
Genotypes:
| Accession | Susceptibility | Aallele | Pedigree | Plant type | Accession source |
| ICGV91114 | Susceptible | A | ICGV 86055' X 'ICGV 86533' | Spanish Bunch | ICRISAT |
| GPBD4 | Resistant | B | KRG 1' X 'CS 16' | Spanish Bunch | inter-specific hybridization |
| TMV2 | Susceptible | A | Mass Selection 'Gudhiatham Bunch' | Spanish Bunch | ICRISAT |
| RBC2F5R12_13 | Resistant | B | Introgression line from 'ICGV 91114' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_15 | Resistant | B | Introgression line from 'ICGV 91114' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_16 | Resistant | B | Introgression line from 'ICGV 91114' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_17 | Resistant | B | Introgression line from 'ICGV 91114' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_18 | Resistant | B | Introgression line from 'ICGV 91114' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_19 | Resistant | B | Introgression line from 'ICGV 91114' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_23 | Resistant | B | Introgression line from 'ICGV 91114' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_25 | Resistant | B | Introgression line from 'ICGV 91114' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_29 | Resistant | B | Introgression line from 'ICGV 91114' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_30 | Resistant | B | Introgression line from 'ICGV 91114' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| JL24 | Susceptible | A | Selection from 'EC 94943' | Spanish Bunch | Oilseeds Research Station, Jalgaon, Maharashtra (India) |
| RBC2F5R12_45 | Resistant | B | Introgression line from 'JL 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_46 | Resistant | B | Introgression line from 'JL 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_78 | Resistant | B | Introgression line from 'JL 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_87 | Resistant | B | Introgression line from 'JL 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_88 | Resistant | B | Introgression line from 'JL 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_97 | Resistant | B | Introgression line from 'JL 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_138 | Resistant | B | Introgression line from 'JL 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_139 | Resistant | B | Introgression line from 'JL 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_140 | Resistant | B | Introgression line from 'JL 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_143 | Resistant | B | Introgression line from 'JL 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| TAG24 | Susceptible | A | TGS2' X 'TGE1' | Spanish Bunch | Bhabha Atomic Research Center, Trombay (India) |
| RBC2F5R12_103 | Resistant | B | Introgression line from 'TAG 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_104 | Resistant | B | Introgression line from 'TAG 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_107 | Resistant | B | Introgression line from 'TAG 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_108 | Resistant | B | Introgression line from 'TAG 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_114 | Resistant | B | Introgression line from 'TAG 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_117 | Resistant | B | Introgression line from 'TAG 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_118 | Resistant | B | Introgression line from 'TAG 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_129 | Resistant | B | Introgression line from 'TAG 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_130 | Resistant | B | Introgression line from 'TAG 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
| RBC2F5R12_133 | Resistant | B | Introgression line from 'TAG 24' X 'GPBD 4' | Parental source 'Spanish Bunch' | Marker-assisted backcrossing |
* aAllele: A = allele present in the rust susceptible genotypes, and B = allele present in the rust resistant genotypes
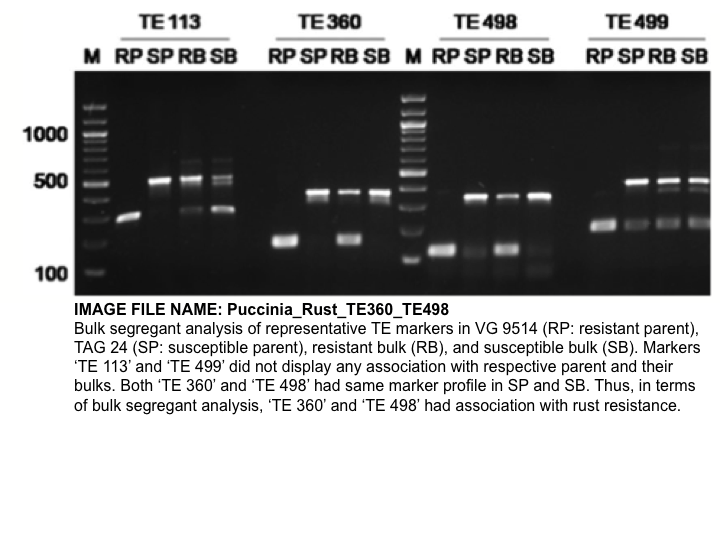
| Marker | TE 360 | TE 498 |
|---|---|---|
| Source | Mondal, Hande et al., 2013 | |
| Comments | "Total of 40 TE primer pairs were found polymorphic between parents and two transposable element markers, and TE 360 and TE 498 were found associated with rust resistance gene. Based on genetic mapping, TE 360 was found linked to the rust resistance gene at 4.5 cM distance." (Mondal, Hande et al., 2013) | |
| Type | Transposable element markers | |
| Repeat | AhMITE1-2 | |
| Forward primer | GGATATGATGCCCATAGCTGA | ATGACTTACATGTAGCAATTG |
| Reverse primer | TGCTGACTACTTGCAATGCC | TGAAAGGAGTCAAAGGTCATG |
| Size (bp) in susceptible lines | approximately 200 | |
| Size (bp) in resistant lines | approximately 450 |
Genotypes:
| Accession | Susceptibility | Aallele | Pedigree | Plant type | Accession source |
| VG 9514 | Resistant | B | CO 1' X 'A. cardenasii' | ||
| TAG 24 | Susceptible | A | TGS2' X 'TGE1' | Spanish Bunch | Bhabha Atomic Research Center, Trombay (India) |
* aAllele: A = allele present in the rust susceptible genotypes, and B = allele present in the rust resistant genotypes
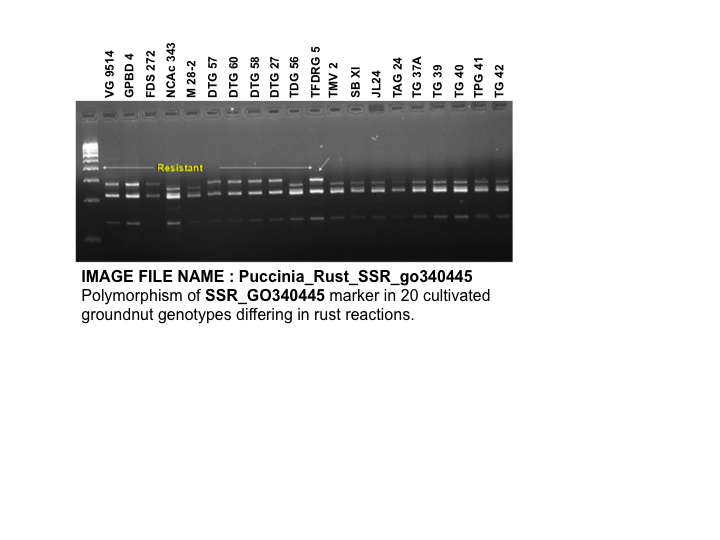
| Marker | SSR_GO340445 | SSR_HO115759 |
|---|---|---|
| Source | Mondel, Badigannavar et al., 2012b | |
| Comments | "Through genetic mapping, EST-SSR markers SSR_ GO340445 and SSR_HO115759 were found closely linked to a rust resistance gene at 1.9 and 3.8 cM distances, respectively." (Mondel, Badigannavar et al., 2012b) | |
| Type | SSR | |
| Repeat | (TC)8 | (GA)9 |
| Forward primer | GGCGGCGGCTGAGGAAGAAG | TATCAACGCAACCTTTTGCAG |
| Reverse primer | ACGCGACGCAGAGTGAAAGAA | GACTTGTGTGGCTGAAACTTGA |
| Size (bp) in susceptible lines | ||
| Size (bp) in resistant lines |
Genotypes:
| Accession | Susceptibility | Aallele | Pedigree | Plant type | Accession source | Comment |
| VG 9514 | Resistant | B | A. cardenasii' X 'CO 1' | NA | NA | Used for making cross |
| GPBD 4 | Resistant | B | KRG 1' X 'ICGV 86855' | Spanish Bunch | NA | Used for original rust resistant screening |
| FDS 272 | Resistant | B | NA | NA | NA | Used for original rust resistant screening |
| NCAc 343 | Resistant | B | NC bunch' X 'PI 1216067' | NA | NA | Used for original rust resistant screening |
| M 28-2 | Resistant | B | EMS mutant of VL 1 | NA | NA | Used for original rust resistant screening |
| DTG 57 | Resistant | B | TAG 24' X 'GPBD 4' | NA | NA | Used for original rust resistant screening |
| DTG 60 | Resistant | B | TG 26' X 'M 28-2' | NA | NA | Used for original rust resistant screening |
| DTG 58 | Resistant | B | TG 26' X 'M 28-2' | NA | NA | Used for original rust resistant screening |
| DTG 27 | Resistant | B | TG 49' X 'B 37c' | NA | NA | Used for original rust resistant screening |
| TDG 56 | Resistant | B | GPBD 4' X 'TG 49' | NA | NA | Used for original rust resistant screening |
| TFDRG 5 | Resistant | B | TAG 24' X 'VG 9514' | NA | NA | Used for original rust resistant screening |
| TMV 2 | Susceptible | B | Mass selection from 'Gudhiatham bunch' | Spanish Bunch | NA | Used for original rust resistant screening |
| SB XI | Susceptible | B | Selection from EC 94943 | NA | NA | Used for original rust resistant screening |
| JL24 | Susceptible | B | Ah 4213' X 'Ah4354' | Spanish Bunch | NA | Used for original rust resistant screening |
| TAG 24 | Susceptible | A | TGS2' X 'TGE1' | Spanish Bunch | Bhabha Atomic Research Center, Trombay (India) | Used for making cross |
| TG 37A | Susceptible | A | TG 25' X 'TG 26' | NA | NA | Used for original rust resistant screening |
| TG 39 | Susceptible | A | TAG 24' X 'TG 19' | NA | NA | Used for original rust resistant screening |
| TG 40 | Susceptible | A | TAG 24' X 'TG 19' | NA | NA | Used for original rust resistant screening |
| TPG 41 | Susceptible | A | TG 28A' X 'TG 22' | NA | NA | Used for original rust resistant screening |
| TG 42 | Susceptible | A | TG 19' X ' TG 26' | NA | NA | Used for original rust resistant screening |
* aAllele: A = allele present in the rust susceptible genotypes, and B = allele present in the rust resistant genotypes
* Accessions 'VG 9514' and 'TAG 24' were used to generate recombinant inbred population. The remaining accessions were originally screened for rust resistance in Mondal and Badigannavar, 2010.
Links:
- * List of Rust, Puccinia QTLs at PeanutBase