Changes between the Arachis hypogaea var. Tifrunner genome assemblies version 1 and version 2
Download files | Change summary table | Chromosome dot plotsOn July 26th, 2019 an improved version of the Arachis hypogaea cv Tifrunner genome was released. Corrections to the A. hypogaea var. Tifrunner genome assembly were made by DNA Zoo, on the basis of Hi-C data produced by the Aiden lab. See also this DNA Zoo announcement.
Downloads
- * Arachis hypogaea var. Tifrunner version 1 pseudomolecules
- * Arachis hypogaea var. Tifrunner version 1 gene models
- * Arachis hypogaea var. Tifrunner version 2 pseudomolecules
- * Arachis hypogaea var. Tifrunner version 2 gene models
- * Arachis hypogaea var. Tifrunner version 1 gene models
A summary of the changes:
- Arahy.01: NO CHANGES
- Arahy.02: Translocation of an 8 Mbp chunk within the pericentromere; swap a bit of the proximal end with Arahy.12
- Arahy.03: Rearrangements of much of the pericentromere; swap a large portion of the distal end with Arahy.13
- Arahy.04: NO CHANGES
- Arahy.05: Swap a bit of the distal end with Arahy.15
- Arahy.06: Swap a large portion of the distal end with Arahy.16
- Arahy.07: Swap a bit of the proximal end with Arahy.17
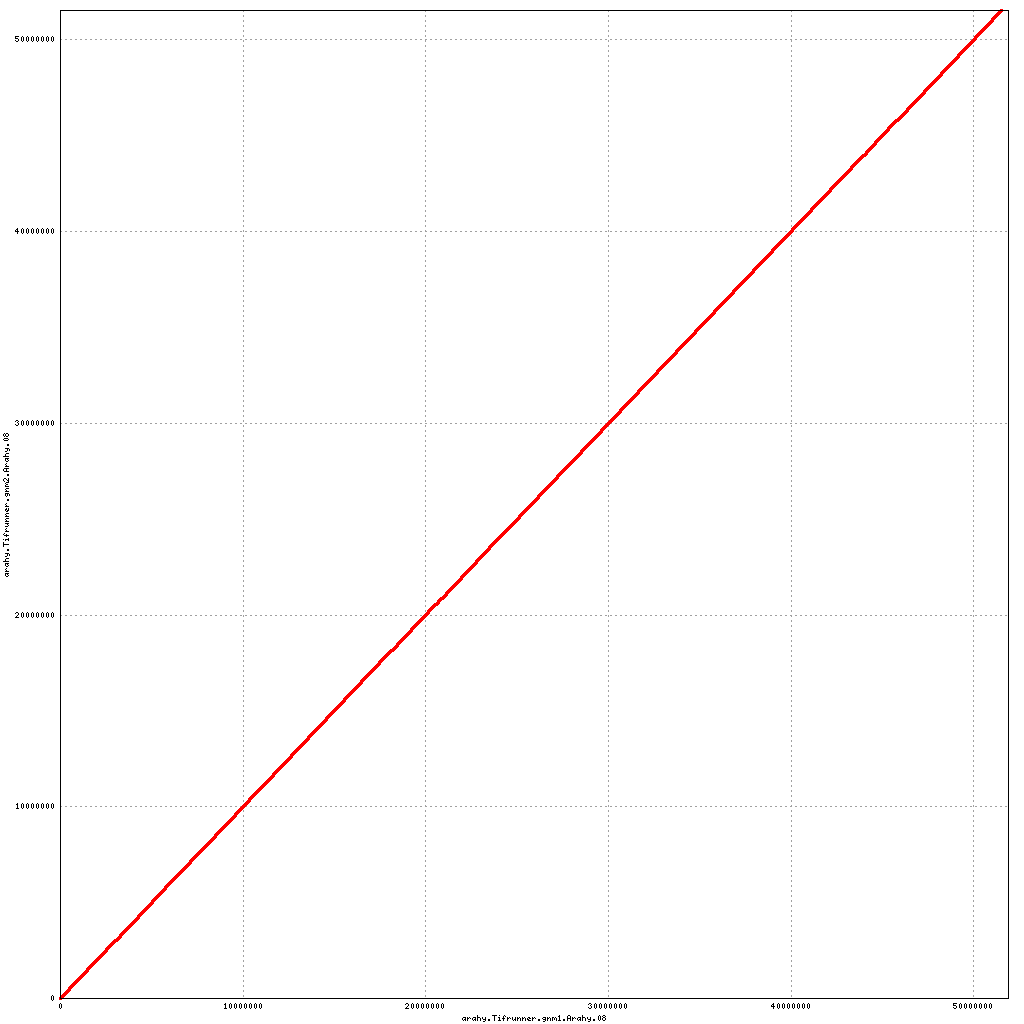
- Arahy.08: Removal of a small scaffold from distal end
- Arahy.09: Inversion of a large centromeric chunk
- Arahy.10: No visible changes
- Arahy.11: No visible changes
- Arahy.12: No visible changes
- Arahy.13: Swap a large portion of the distal end with Arahy.03
- Arahy.14: NO CHANGES
- Arahy.15: Inversion of a large centromeric chunk
- Arahy.16: Swap a large portion of the distal end with Arahy.06
- Arahy.17: Swap a bit of the proximal end with Arahy.07
- Arahy.18: No visible changes
- Arahy.19: Addition of a small (700 kb) scaffold to proximal end
- Arahy.20: Addition of a small (1 Mb) scaffold to distal end
- Arahy.02: Translocation of an 8 Mbp chunk within the pericentromere; swap a bit of the proximal end with Arahy.12
Images of changes by chromosome
(Produced by MUMmer)
Arahy.01 |
Arahy.02 |
Arahy.03 |
Arahy.04 |
Arahy.05 |
Arahy.06 |
Arahy.07 |
Arahy.08 |
Arahy.09 |
Arahy.10
Arahy.11 |
Arahy.12 |
Arahy.13 |
Arahy.14 |
Arahy.15 |
Arahy.16 |
Arahy.17 |
Arahy.18 |
Arahy.19 |
Arahy.20
Pseudomolecule Arahy.01 x Arahy.01 (X-axis=gnm1, Y-axis=gnm2)
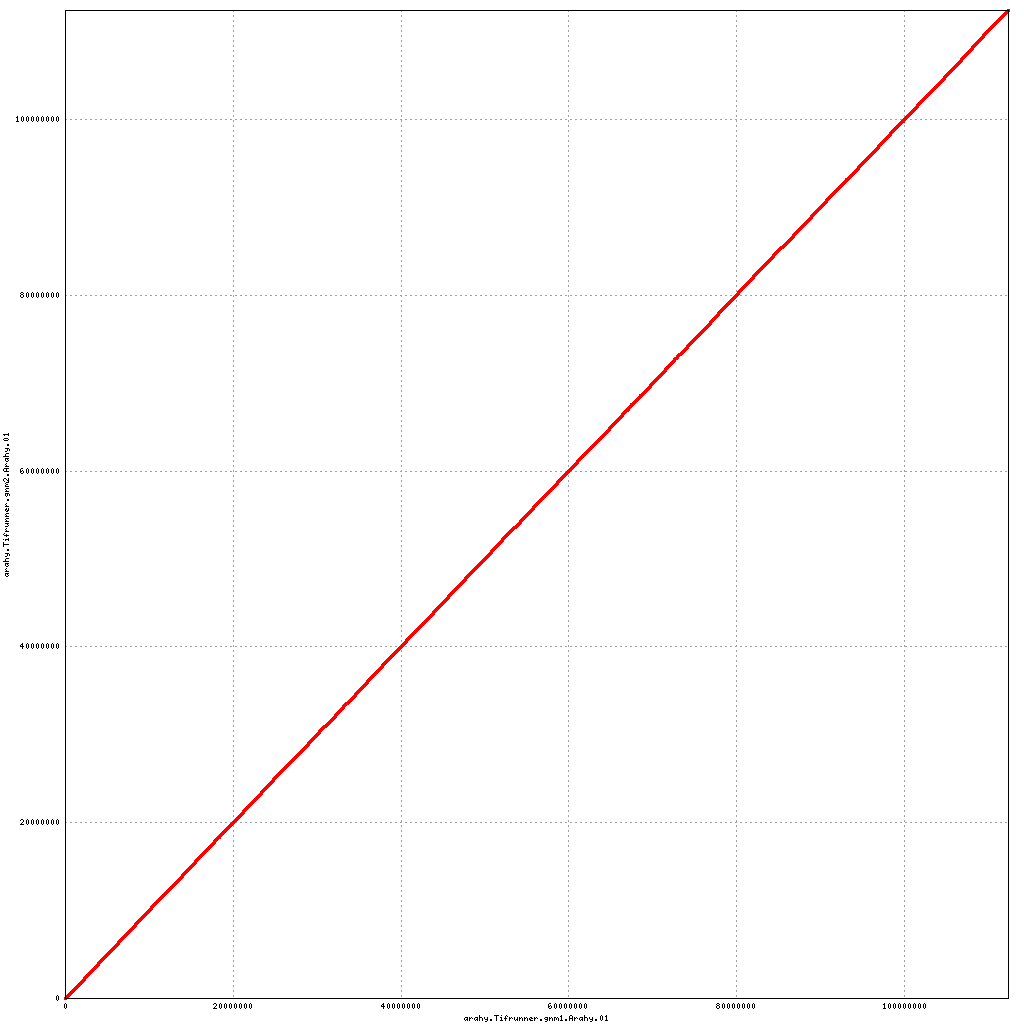
top
Pseudomolecule Arahy.02 x Arahy.02 (X-axis=gnm1, Y-axis=gnm2)
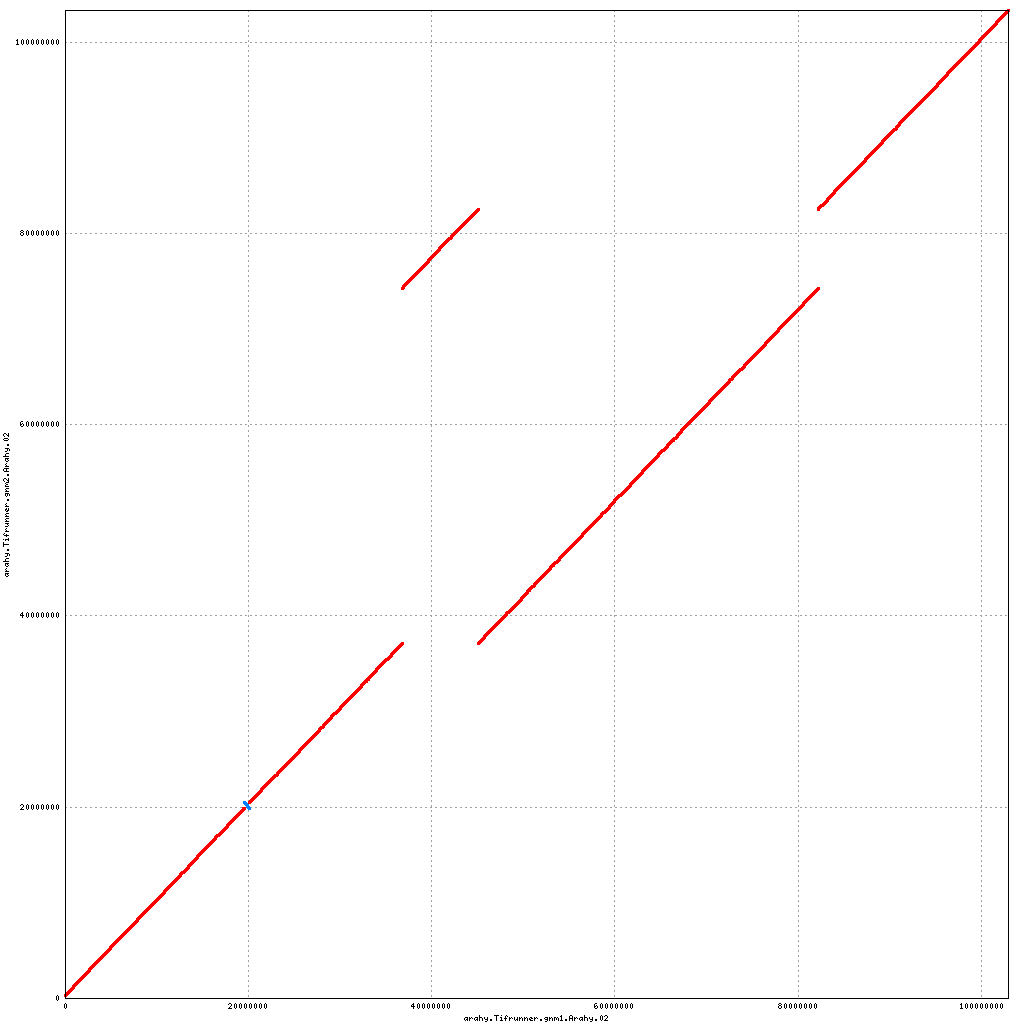
top
Pseudomolecule Arahy.03 x Arahy.03 (X-axis=gnm1, Y-axis=gnm2)
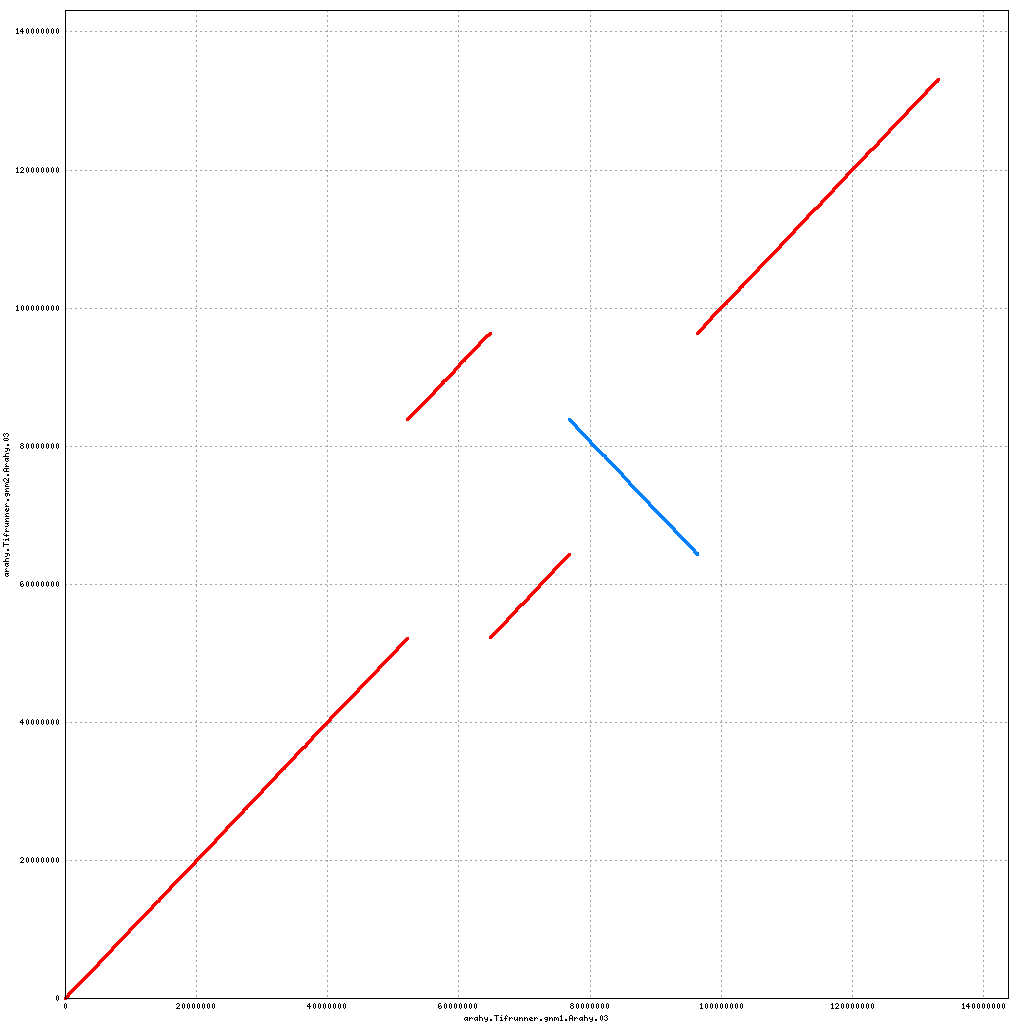
Pseudomolecule Arahy.03 x Arahy.13 (X-axis=gnm1, Y-axis=gnm2)
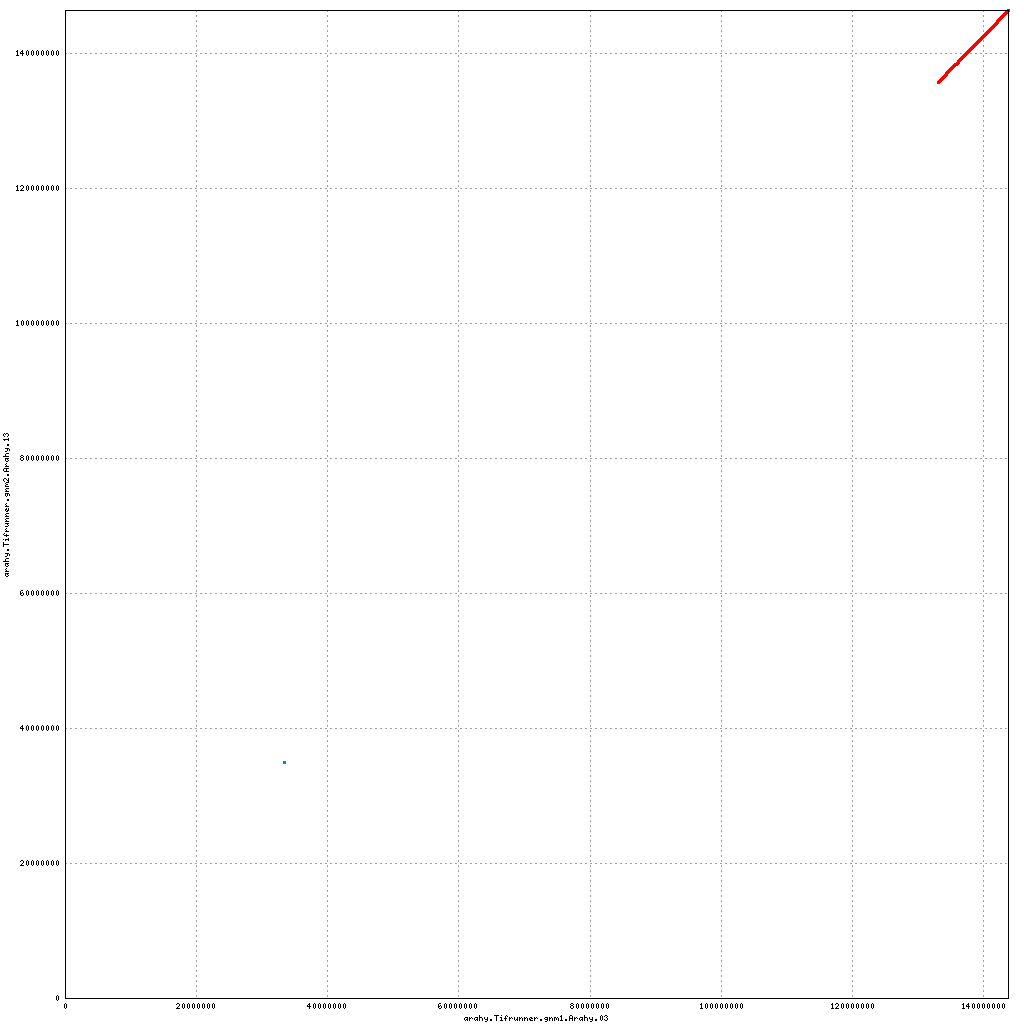
top
Pseudomolecule Arahy.04 x Arahy.04 (X-axis=gnm1, Y-axis=gnm2)
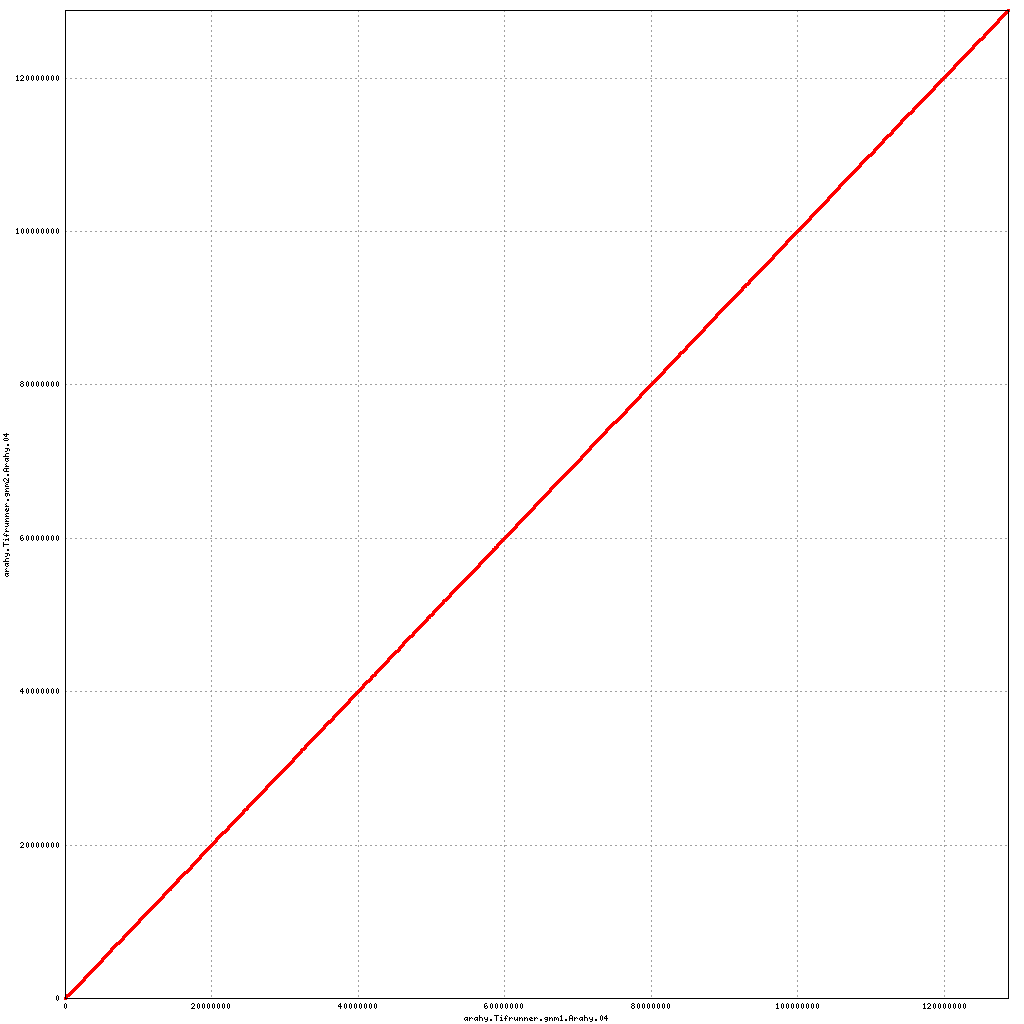
top
Pseudomolecule Arahy.05 x Arahy.05 (X-axis=gnm1, Y-axis=gnm2)
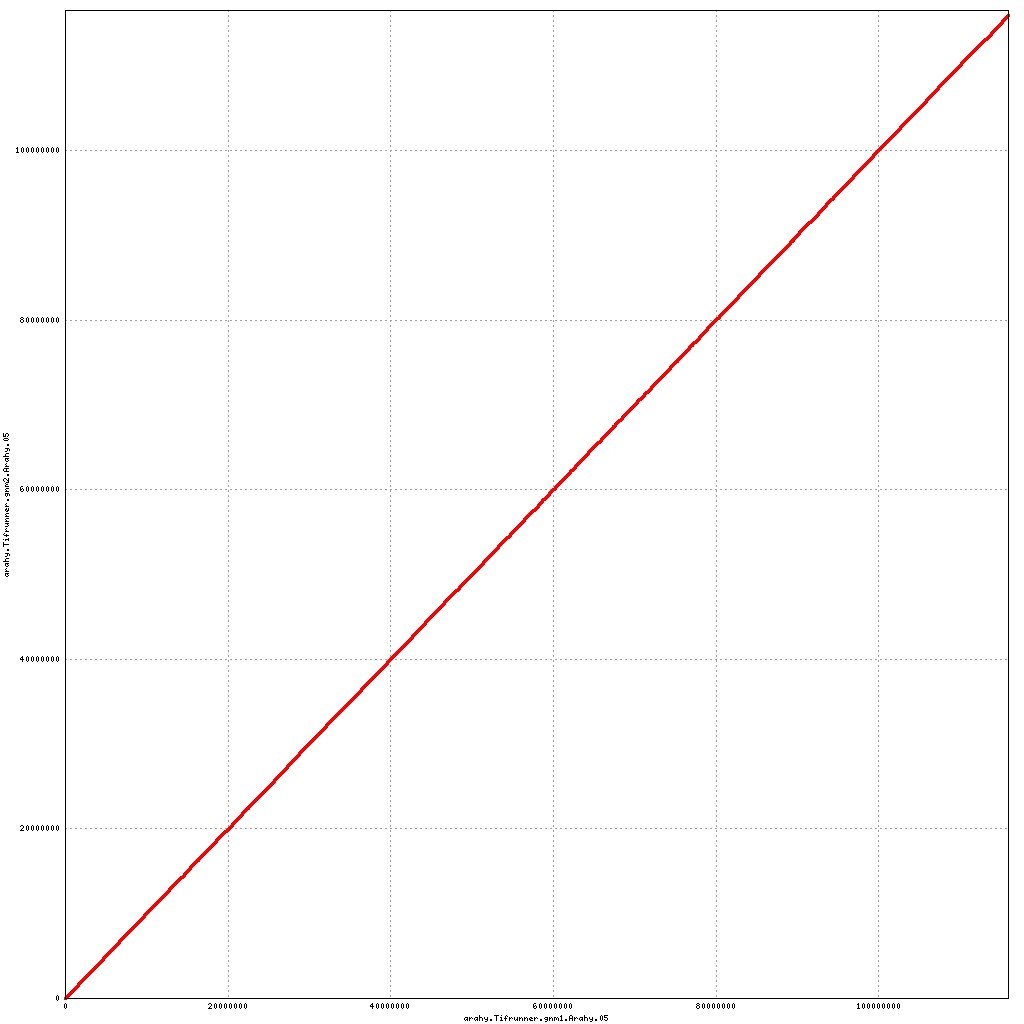
top
Pseudomolecule Arahy.06 x Arahy.06 (X-axis=gnm1, Y-axis=gnm2)
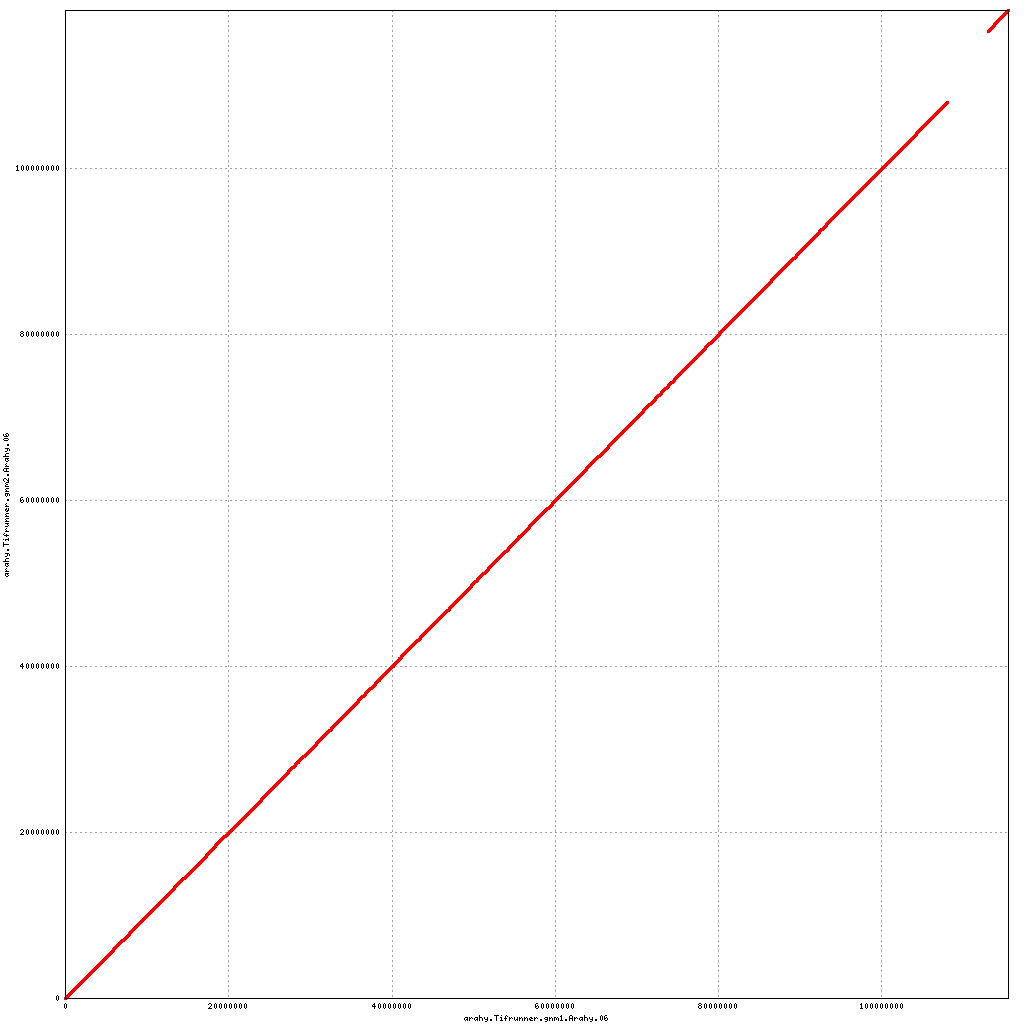
Pseudomolecule Arahy.06 x Arahy.16 (X-axis=gnm1, Y-axis=gnm2)
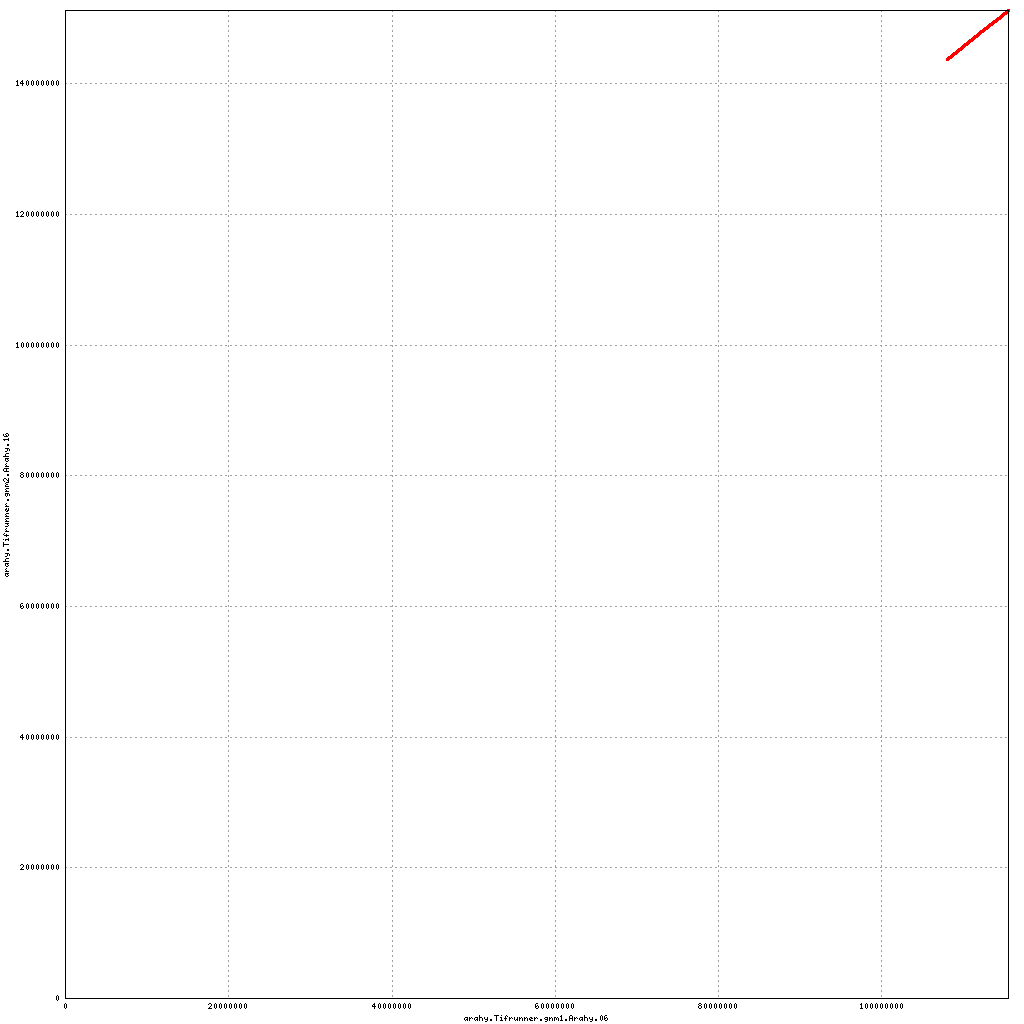
top
Pseudomolecule Arahy.07 x Arahy.07 (X-axis=gnm1, Y-axis=gnm2)
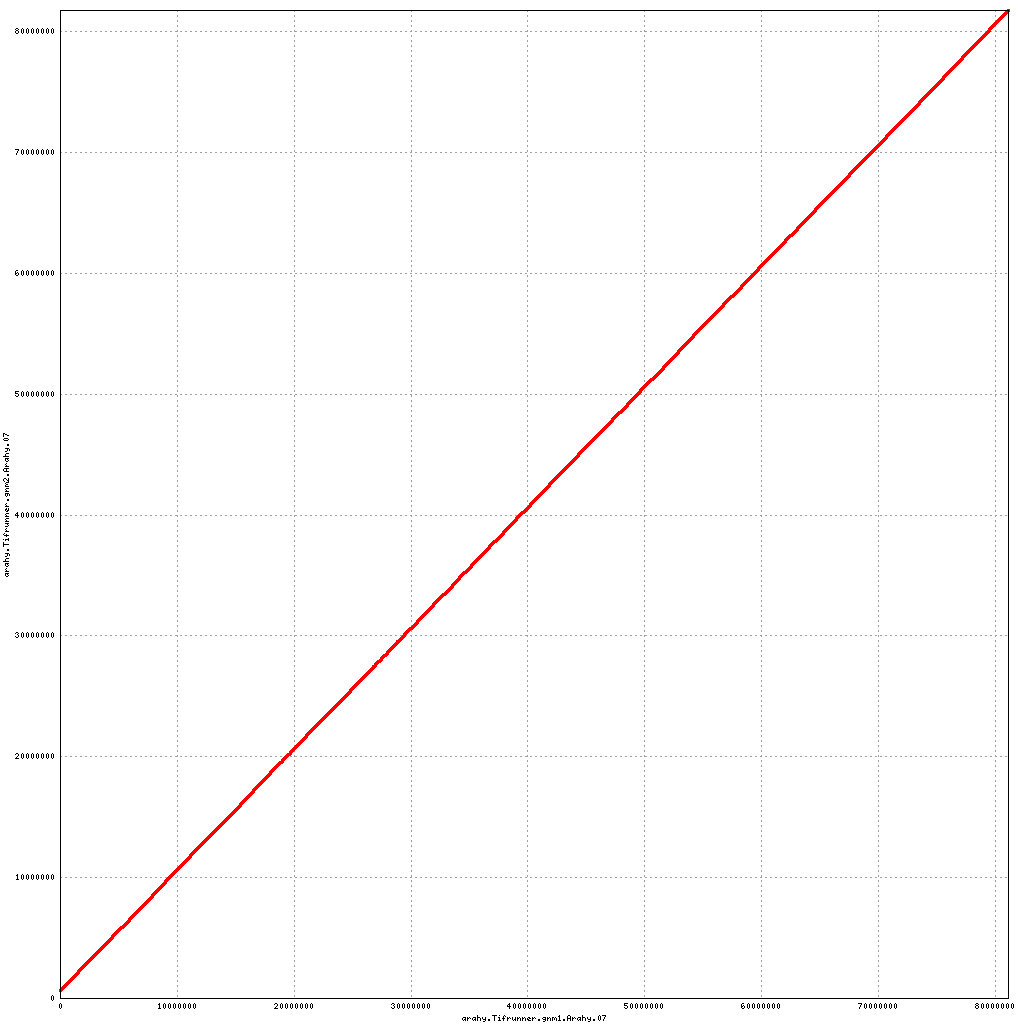
top
Pseudomolecule Arahy.08 x Arahy.08 (X-axis=gnm1, Y-axis=gnm2)
Pseudomolecule Arahy.08 x Arahy.02 (X-axis=gnm1, Y-axis=gnm2)
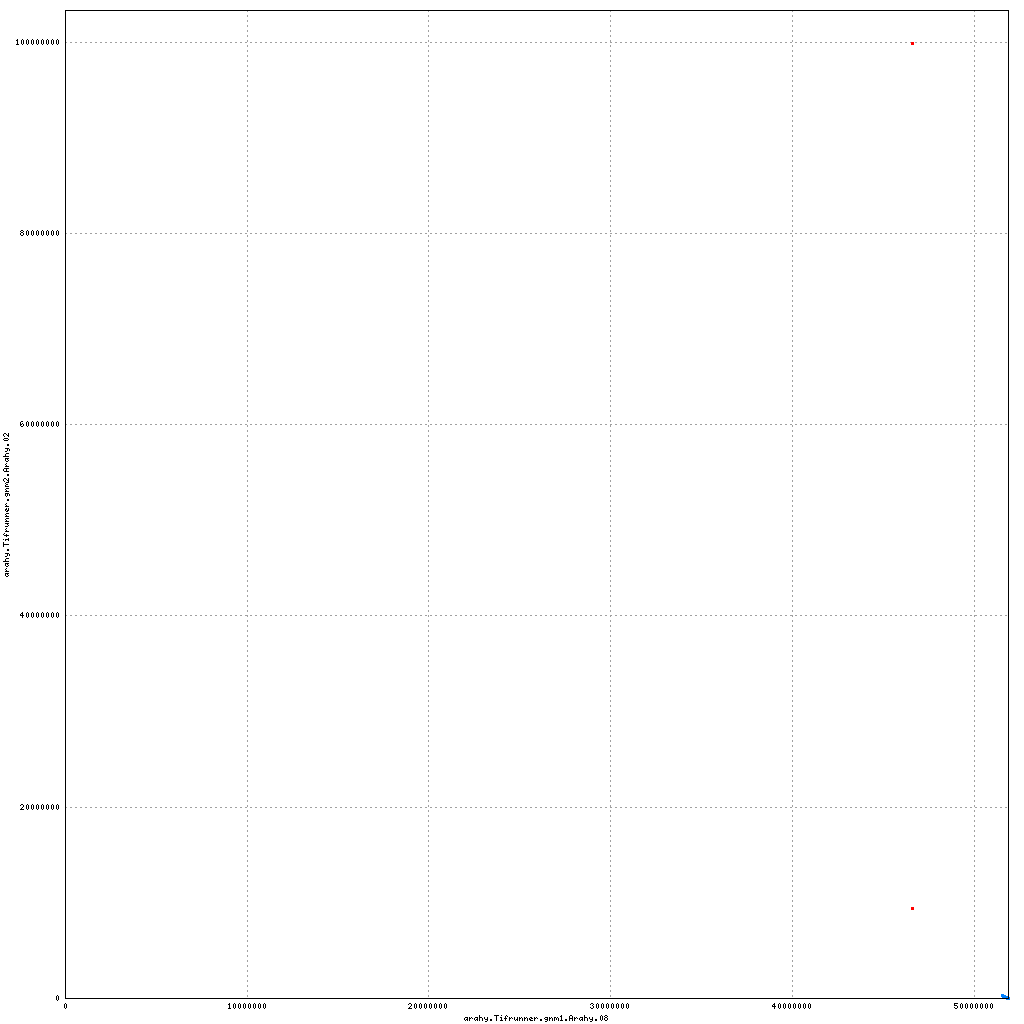
top
Pseudomolecule Arahy.09 x Arahy.09 (X-axis=gnm1, Y-axis=gnm2)
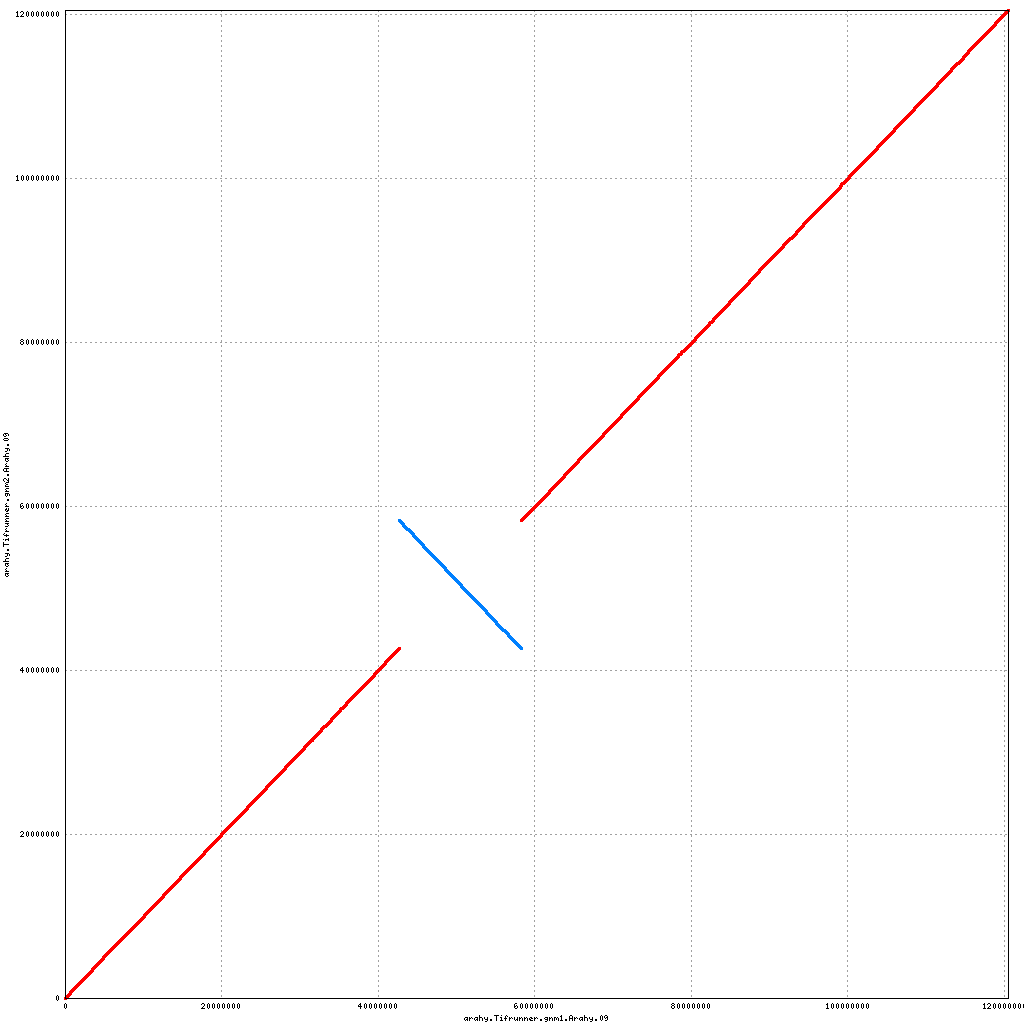
top
Pseudomolecule Arahy.10 x Arahy.10 (X-axis=gnm1, Y-axis=gnm2)
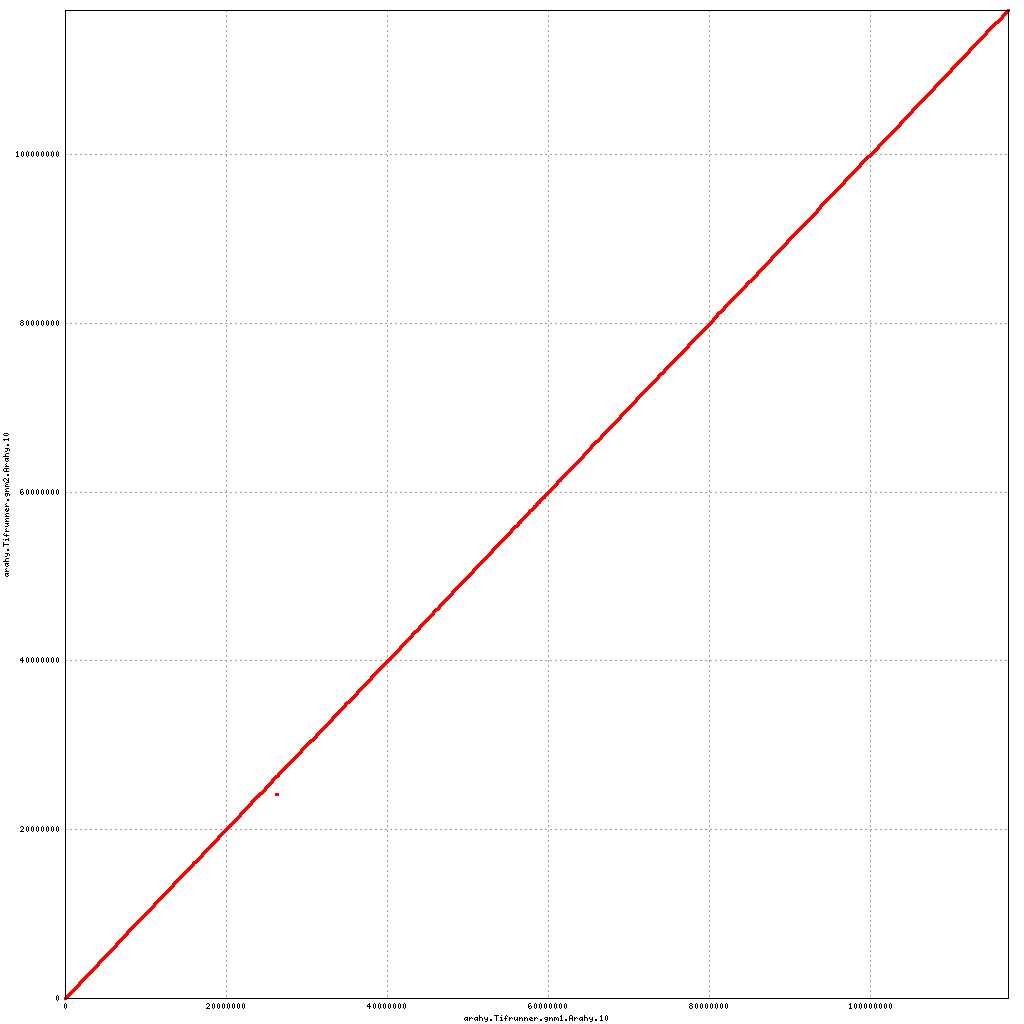
top
Pseudomolecule Arahy.11 x Arahy.11 (X-axis=gnm1, Y-axis=gnm2)
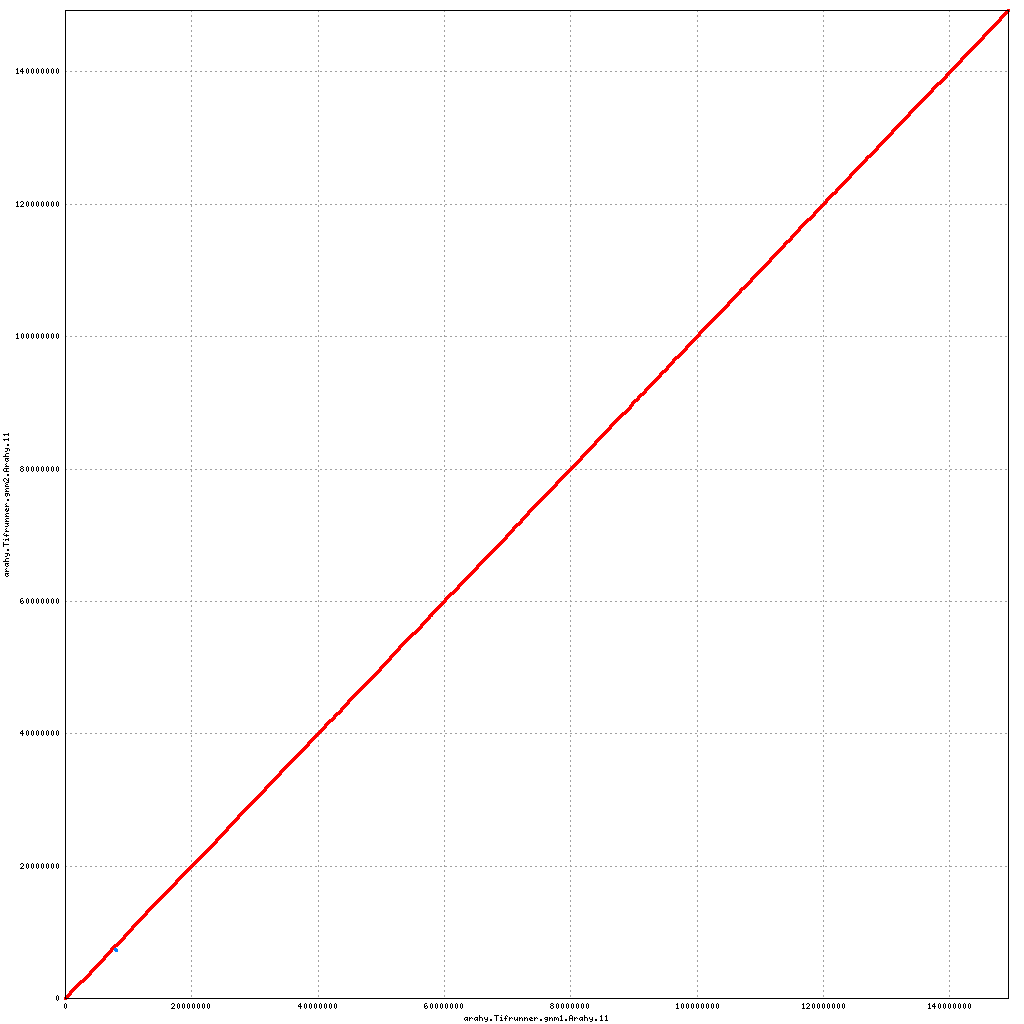
top
Pseudomolecule Arahy.12 x Arahy.12 (X-axis=gnm1, Y-axis=gnm2)
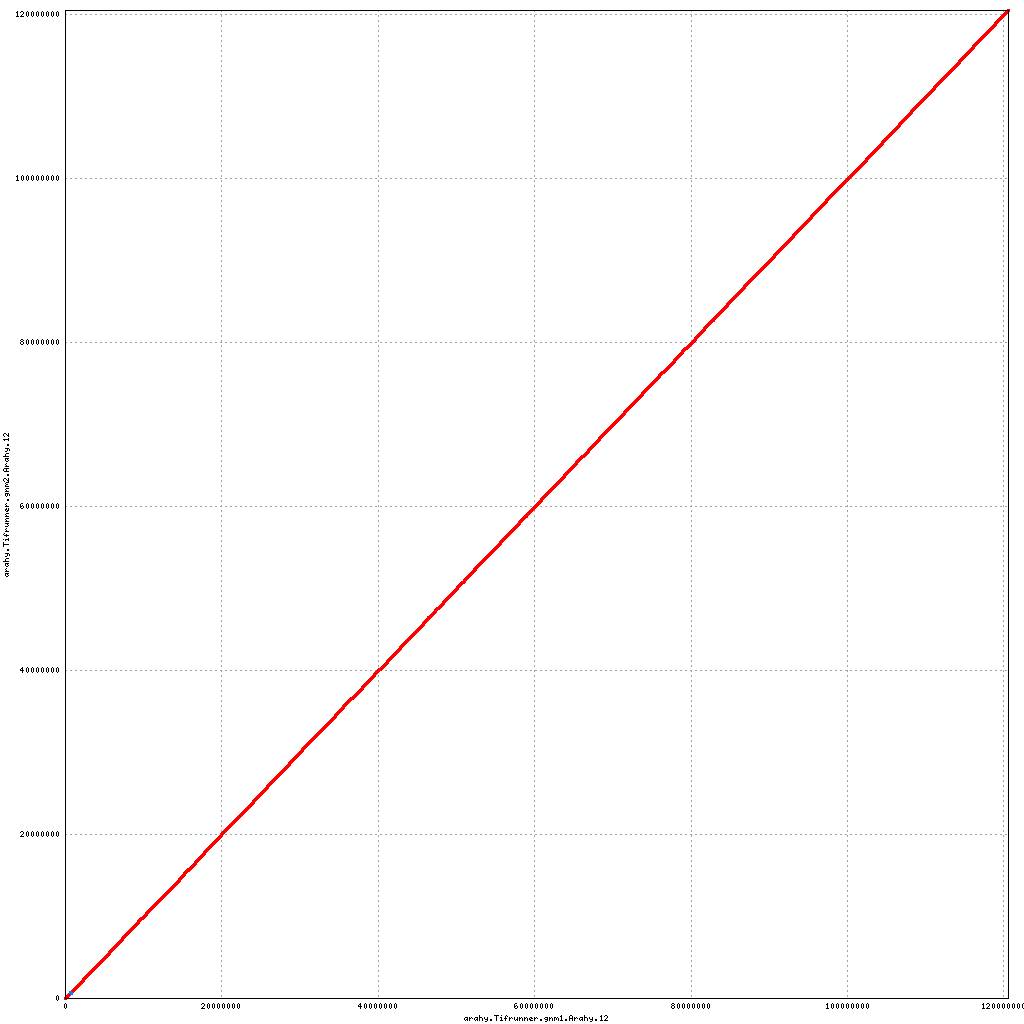
Pseudomolecule Arahy.12 x Arahy.05 (X-axis=gnm1, Y-axis=gnm2)
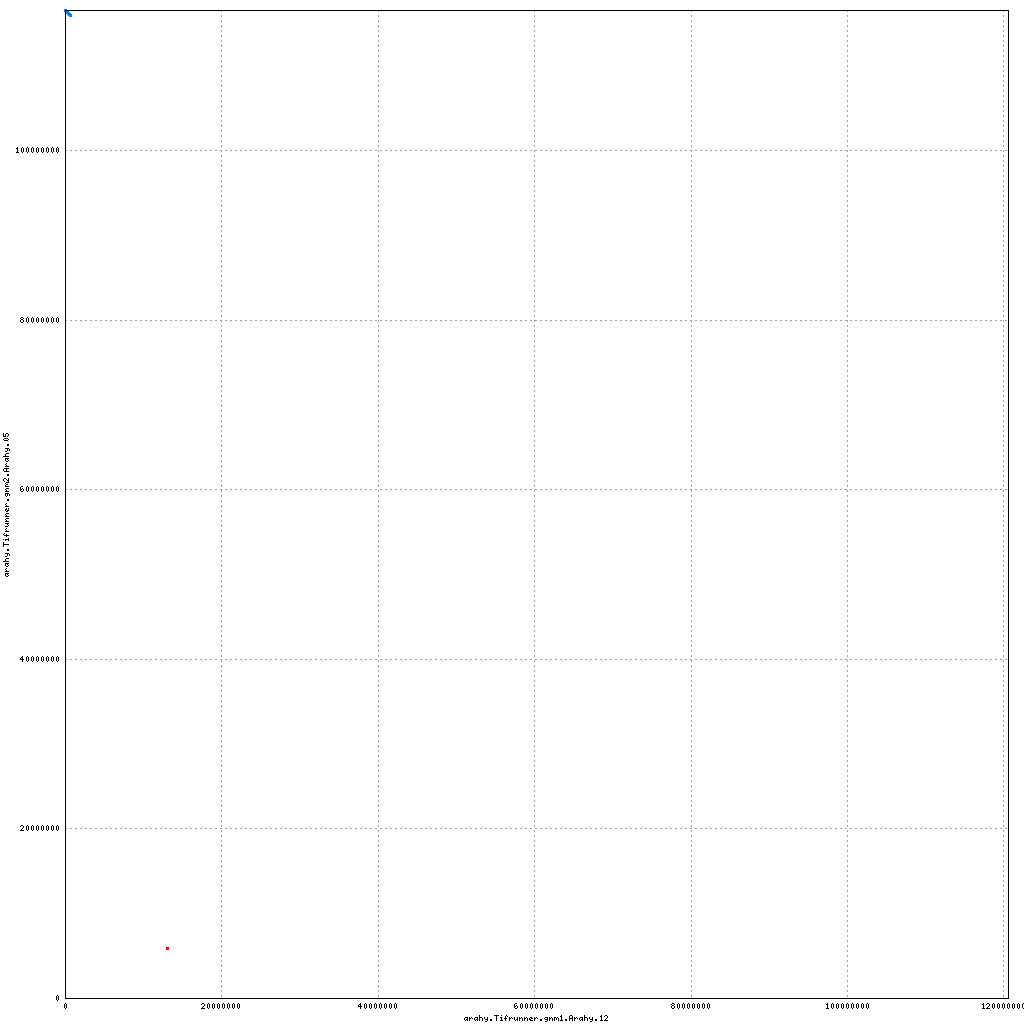
top
Pseudomolecule Arahy.13 x Arahy.13 (X-axis=gnm1, Y-axis=gnm2)
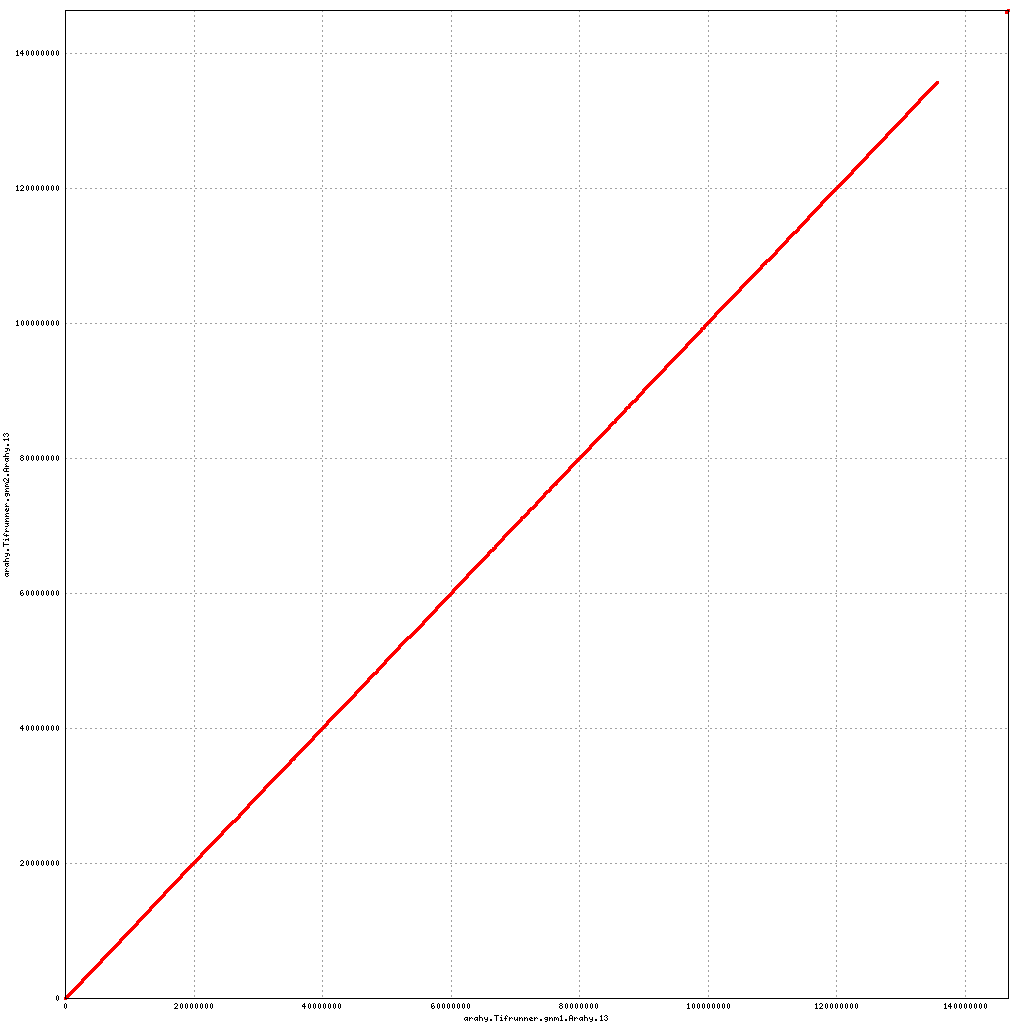
Pseudomolecule Arahy.13 x Arahy.03 (X-axis=gnm1, Y-axis=gnm2)
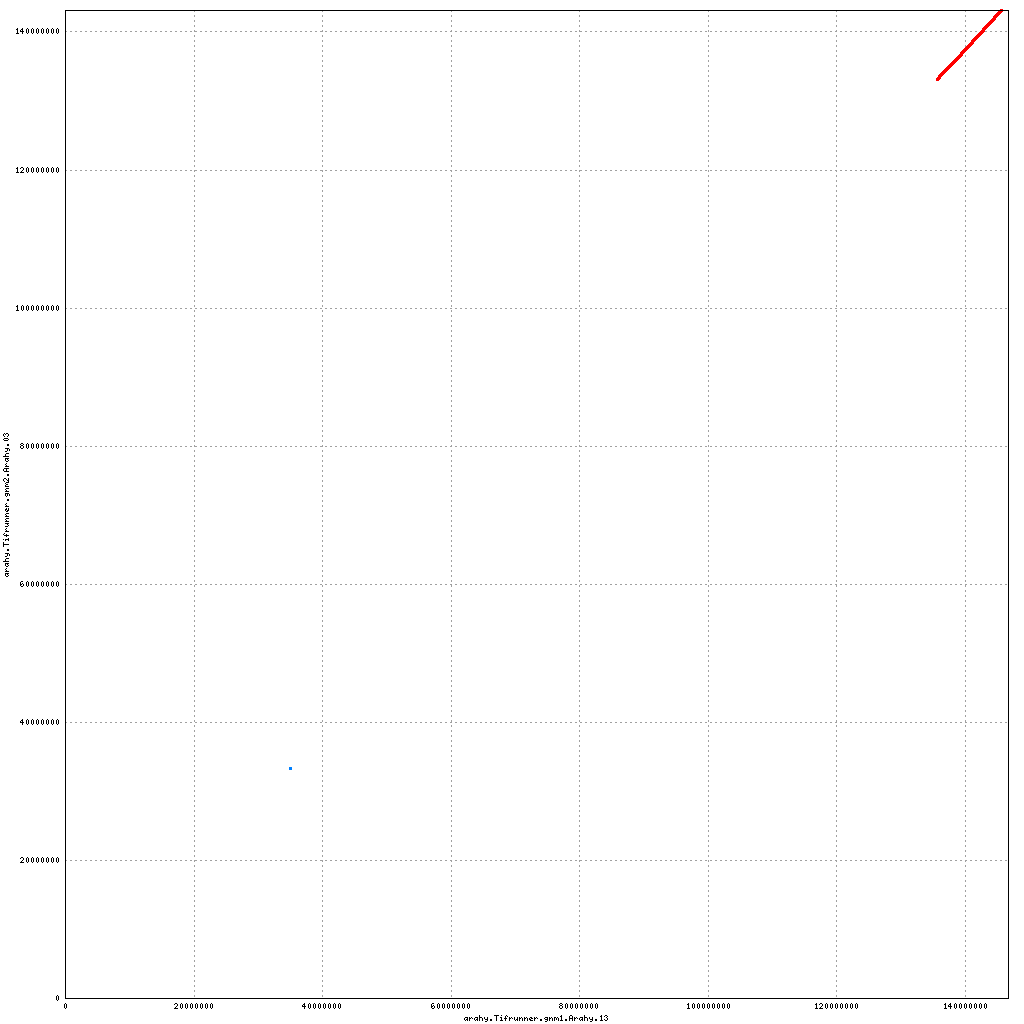
Pseudomolecule Arahy.13 x Arahy.20 (X-axis=gnm1, Y-axis=gnm2)
top
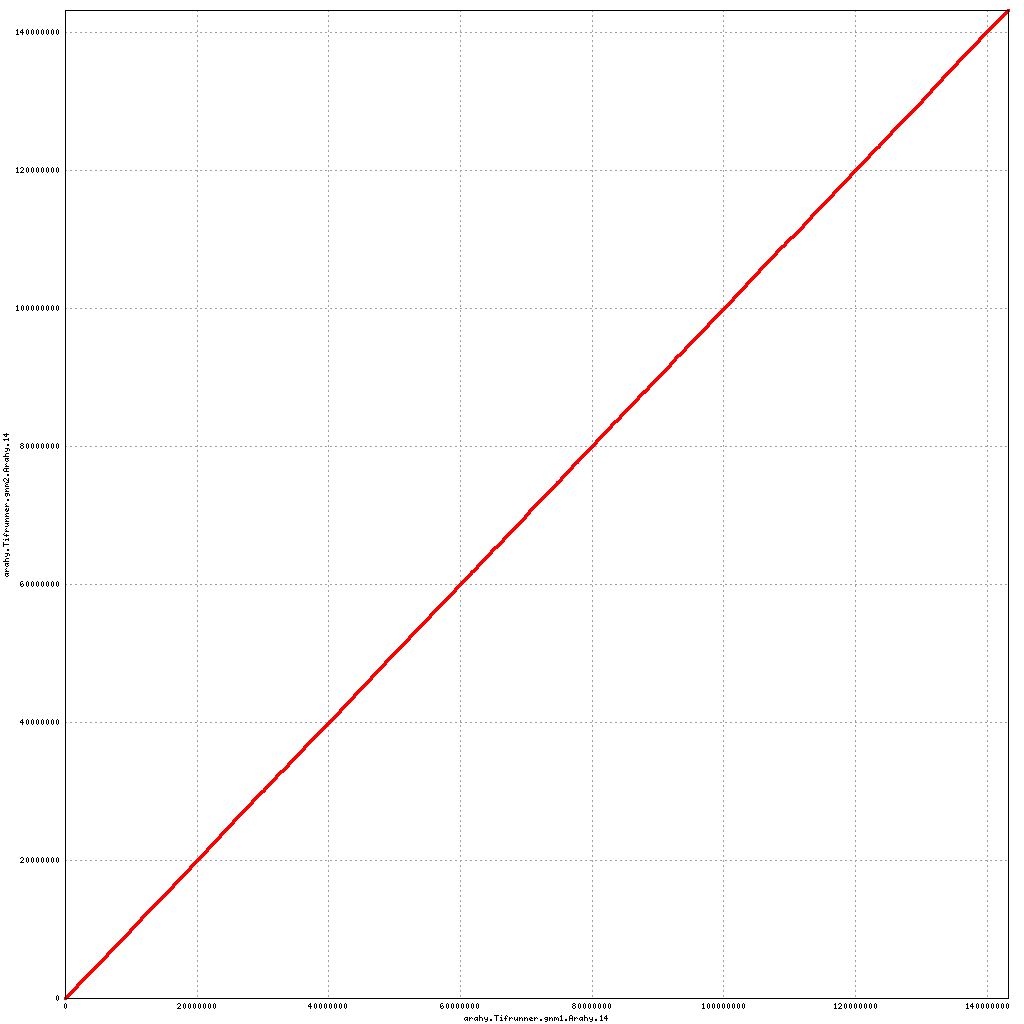
Pseudomolecule Arahy.14 x Arahy.14 (X-axis=gnm1, Y-axis=gnm2)
top
Pseudomolecule Arahy.15 x Arahy.15 (X-axis=gnm1, Y-axis=gnm2)
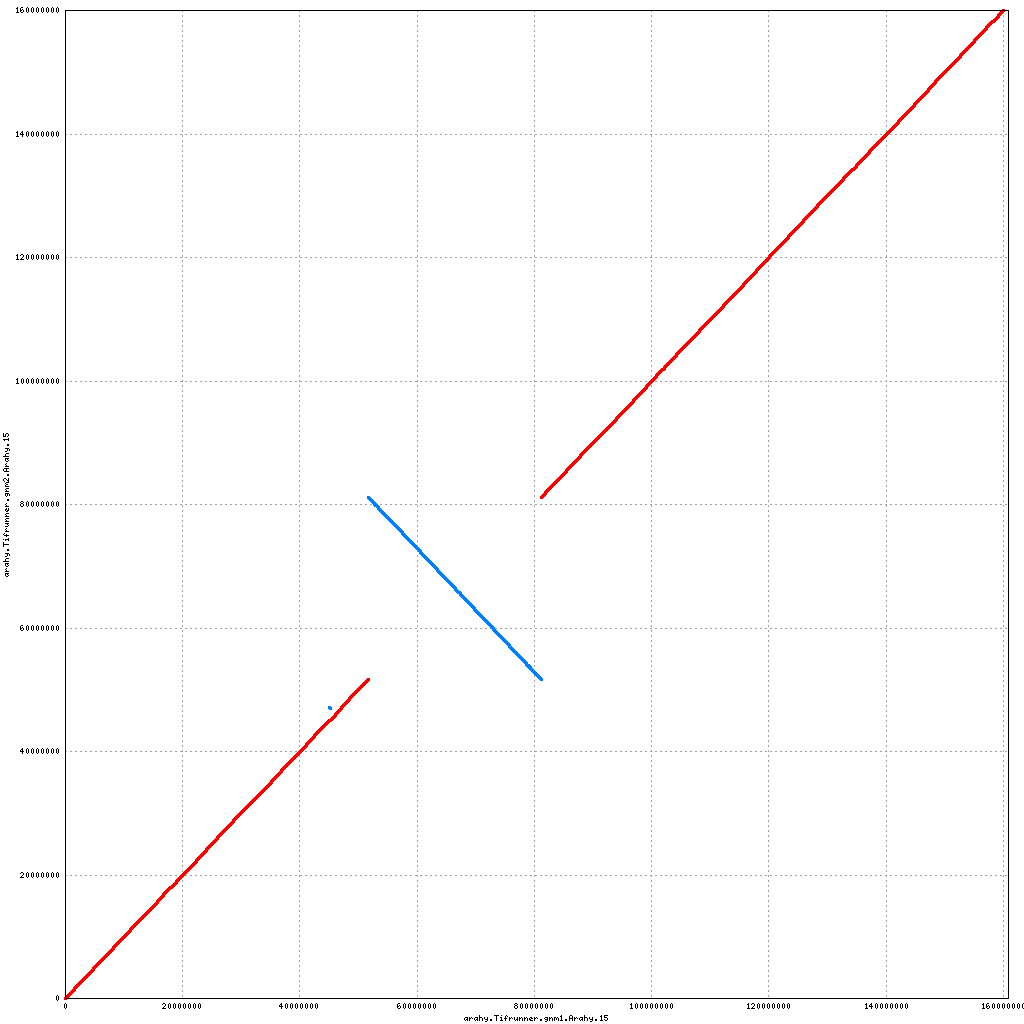
Pseudomolecule Arahy.15 x Arahy.19 (X-axis=gnm1, Y-axis=gnm2)
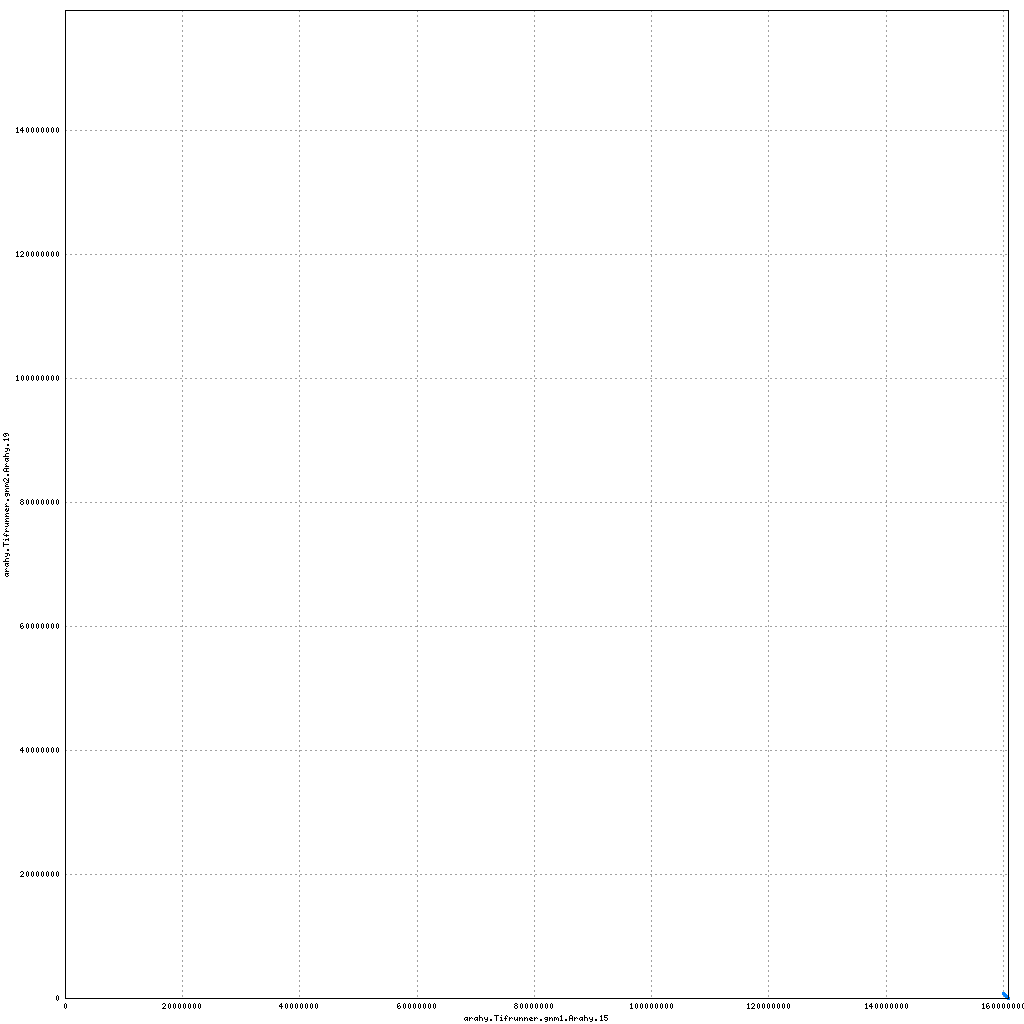
top
Pseudomolecule Arahy.16 x Arahy.16 (X-axis=gnm1, Y-axis=gnm2)
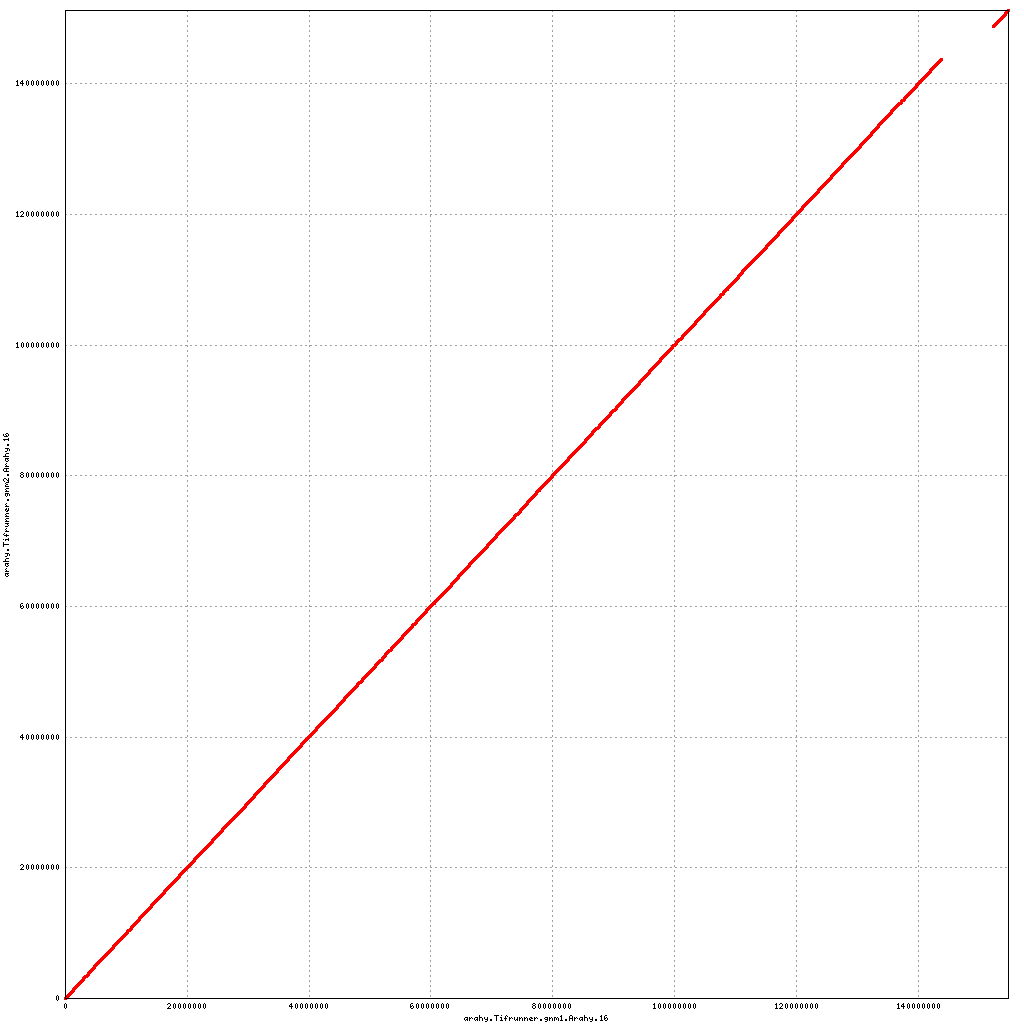
Pseudomolecule Arahy.16 x Arahy.06 (X-axis=gnm1, Y-axis=gnm2)

top
Pseudomolecule Arahy.17 x Arahy.17 (X-axis=gnm1, Y-axis=gnm2)
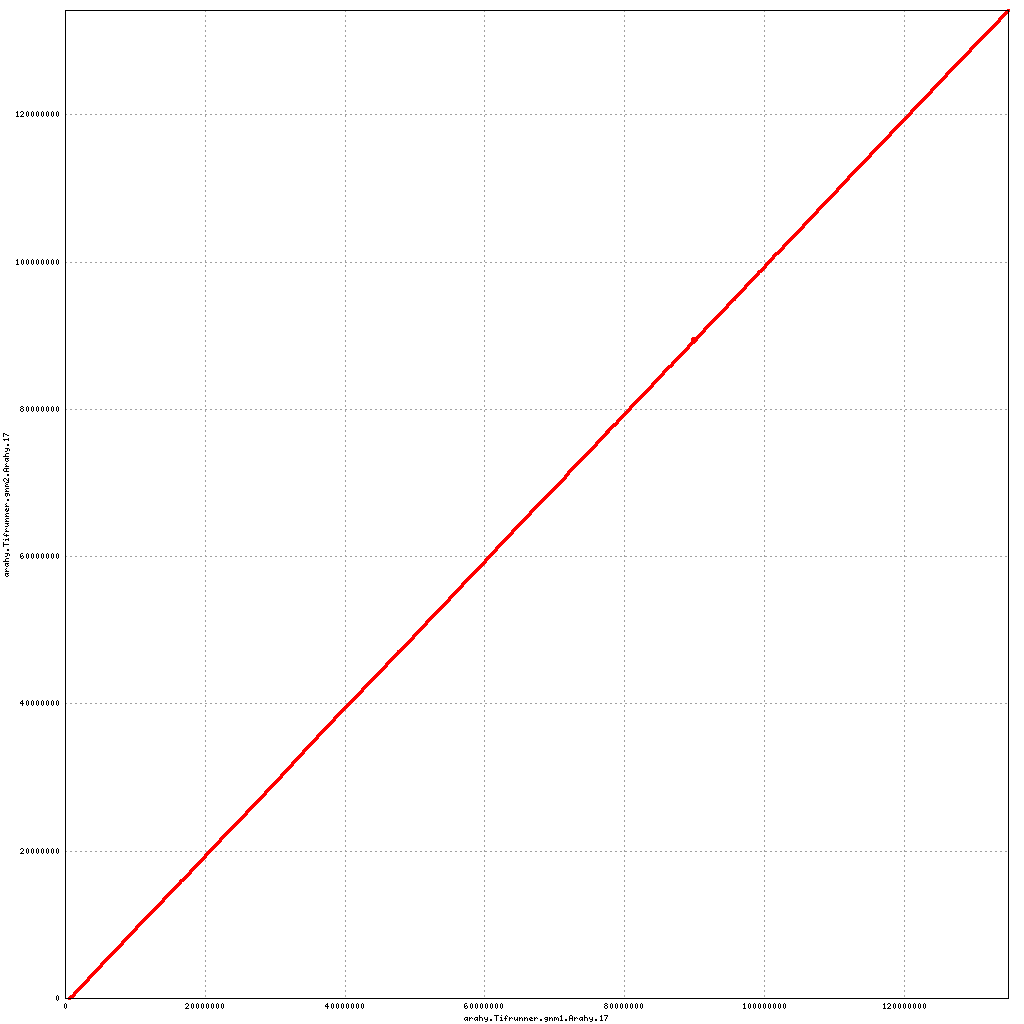
Pseudomolecule Arahy.17 x Arahy.07 (X-axis=gnm1, Y-axis=gnm2)
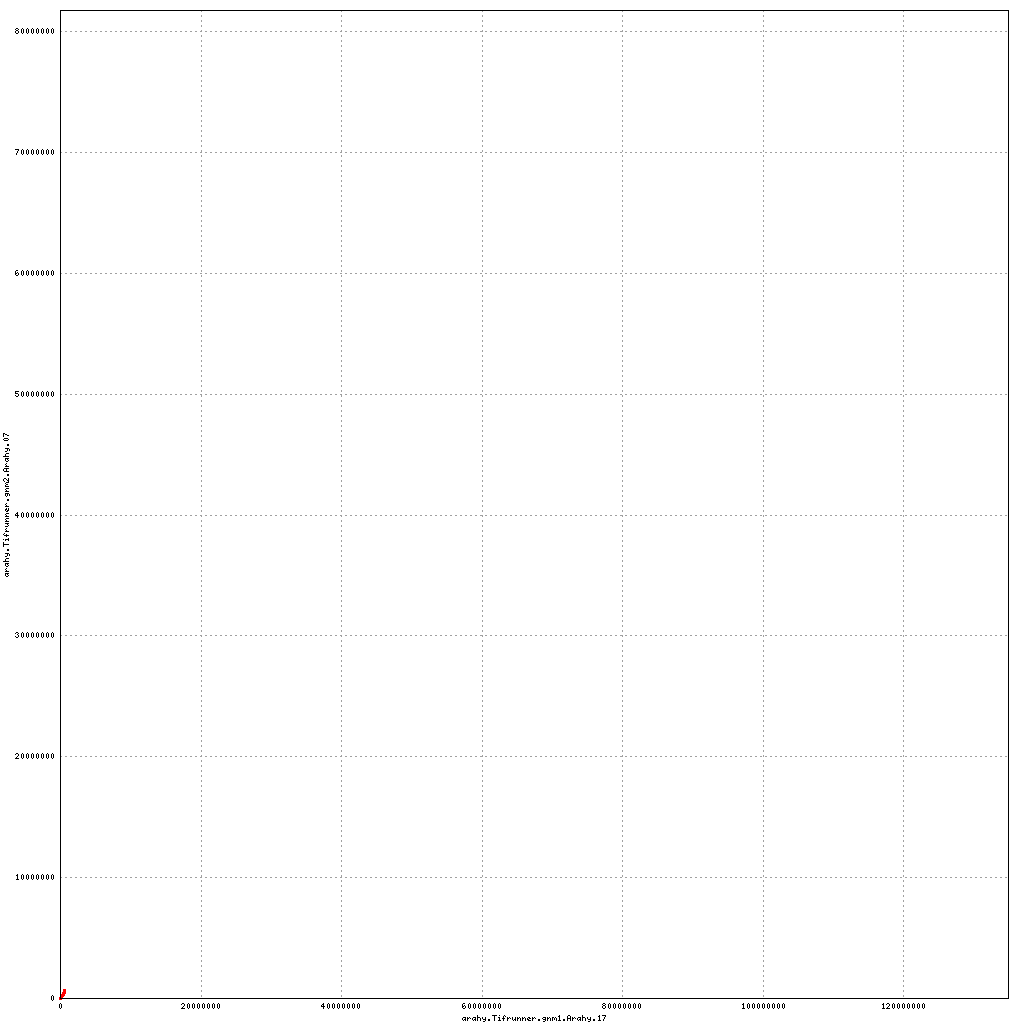
top
Pseudomolecule Arahy.18 x Arahy.18 (X-axis=gnm1, Y-axis=gnm2)
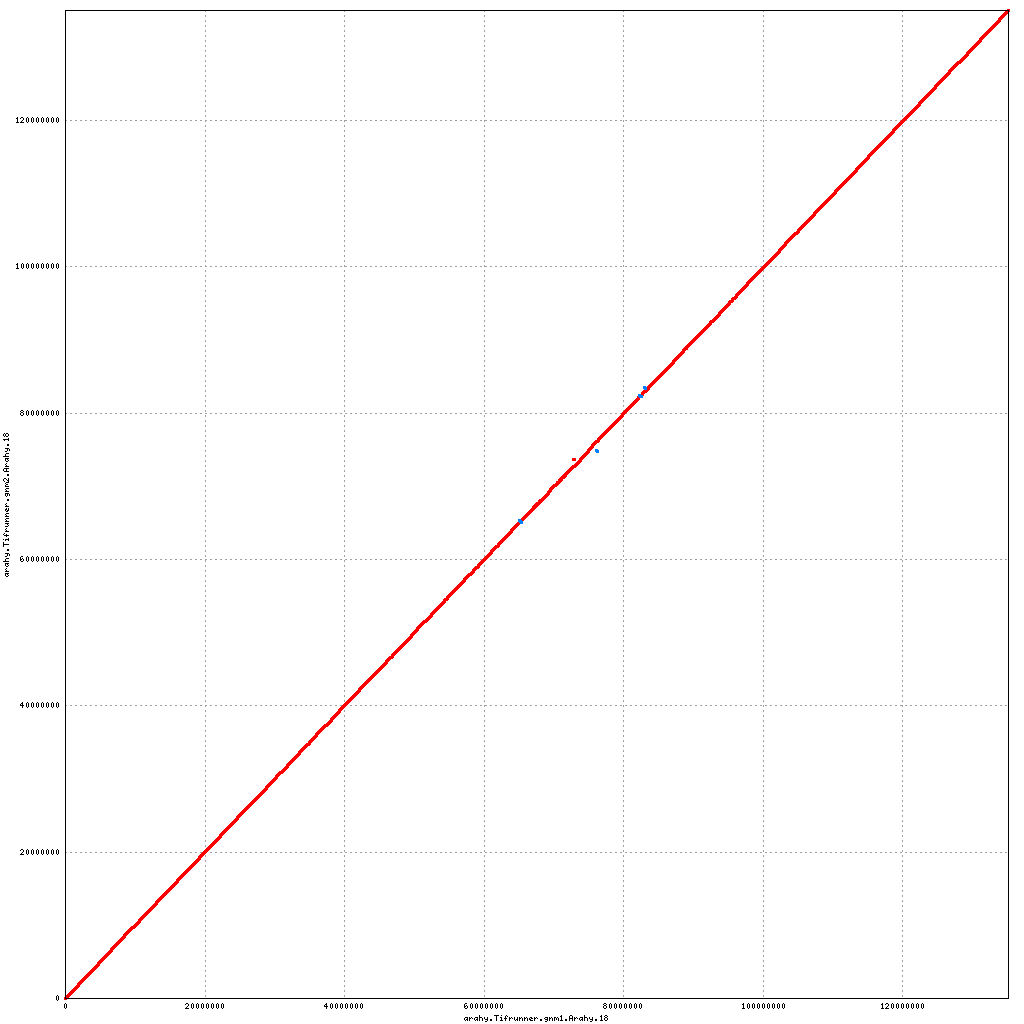
top
Pseudomolecule Arahy.19 x Arahy.19 (X-axis=gnm1, Y-axis=gnm2)
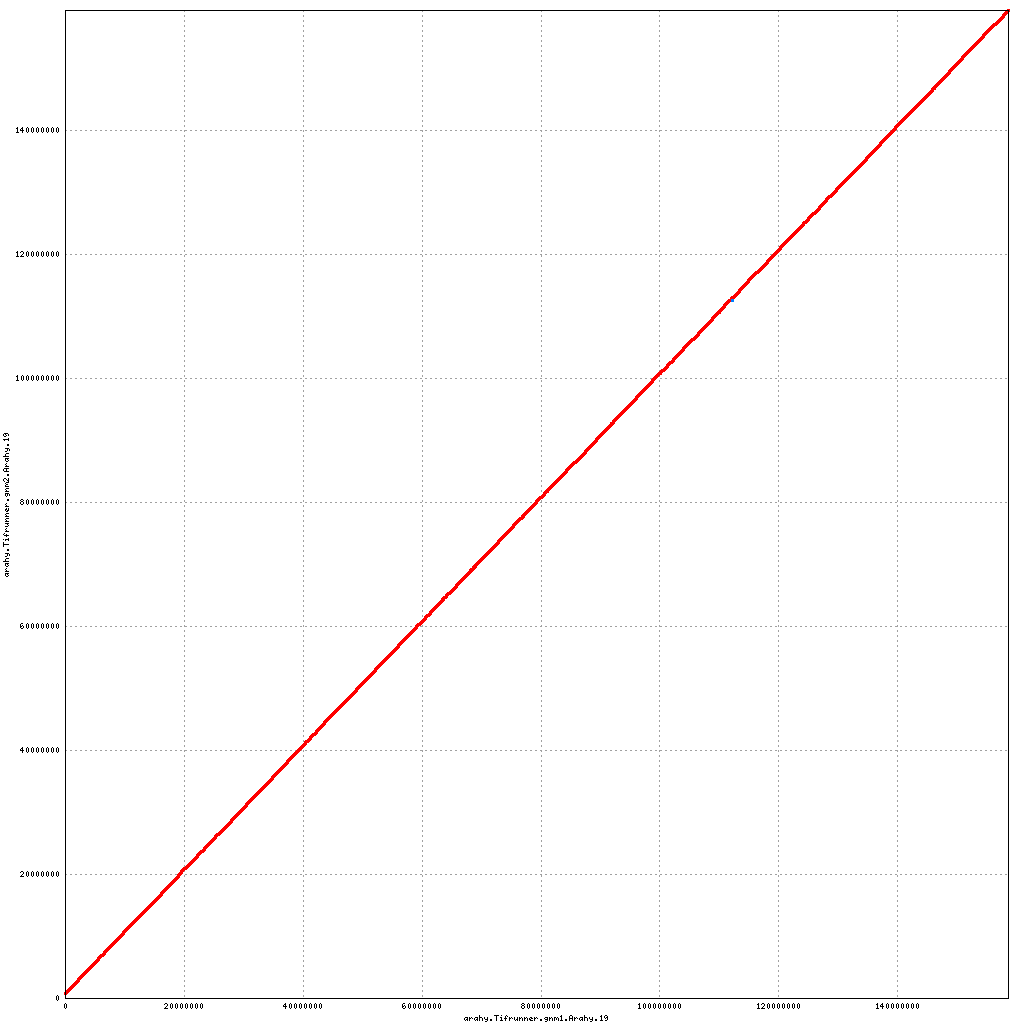
top
Pseudomolecule Arahy.20 x Arahy.20 (X-axis=gnm1, Y-axis=gnm2)
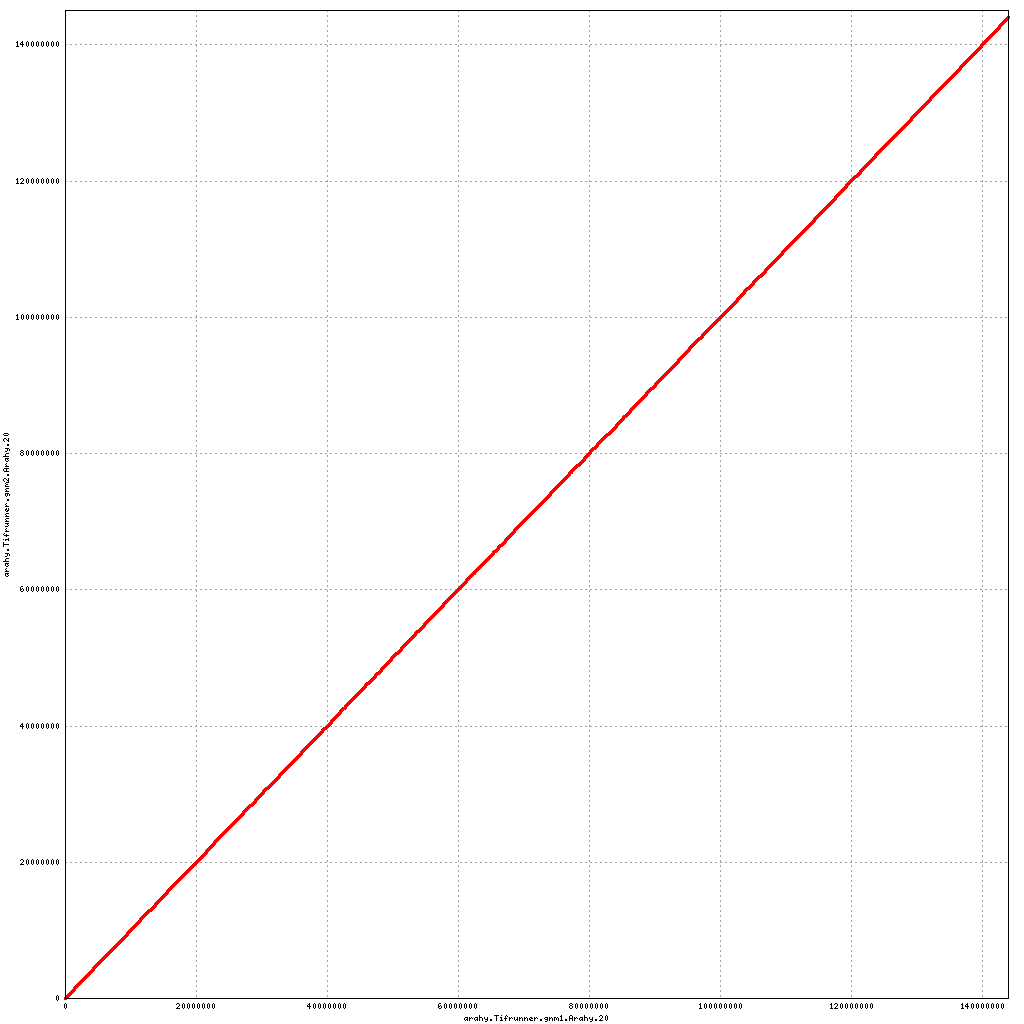
top